Cassette Exon Finder
Finding cassette exon based on gene annotation
This project is maintained by Puriney
Alternative Splicing & Cassette Exon
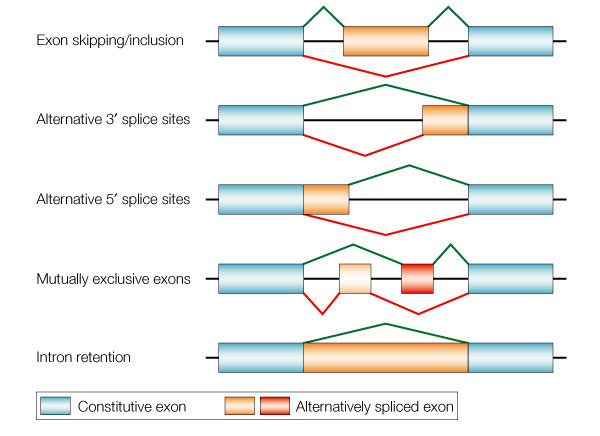
Alternative splicing makes main contributions to mamaalian trancriptome dynamics, and futher proteomic complexity. Cassette exon is the largest category (over 60%) of alternative exon which includes many other forms, like alternative 5' splicing, alternative 3' splicing, mutually exlusive exon, etc. [1,2] Cassette exon is related to disease, like SMA (Spinal Muscular Atrophy), a neorodegenerative disorder, which is direclty caused by SMN1 defiency. The inclusion of exon-7 among SMN gene leads to SMN1 transcripts in normal people, while the unexpected exlusion (or skipping) leads to SMN2 which finally results in infantile death. [3,4] This example indicates RNA splicing is worthwhile to be studied and also that is why SMN1/SMN2 becomes the model isoform in RNA splicing research field.
Work Flow
This package aims to find all the cassette exon based on well-documented annotation. Indeed, this is simplly texts filering work.

The key is to find all the exons absent in one isoform are precisely present in the corresponding intron of anther differnt isoform among the same gene.

Perl Scripts
uniq_intron_producer.plexon_with_flanking_corrd_producer.plstrict_cassette_exon_producer.pl-
cassette_exon_simplifier.pl(which is optional)
Others
As Perl is used as a scaffold, BEDTools toolbox must be employed here for convinience.
Refs
- Wang, E. T., Sandberg, R., Luo, S., Khrebtukova, I., Zhang, L., Mayr, C., Kingsmore, S. F., et al. (2008). Alternative isoform regulation in human tissue transcriptomes. Nature.
- Cartegni, L., Chew, S. L., & Krainer, A. R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nature Reviews Genetics 3, 285–298 (2002)
- Scholl, R., Meyer, K., & Schümperli, D. (2007). Position and functional importance of the branch site preceding SMN exon7, (March), 34–37.
- Coady, T. H., Baughan, T. D., Shababi, M., Passini, M. a, & Lorson, C. L. (2008). Development of a single vector system that enhances trans-splicing of SMN2 transcripts. PloS one, 3(10), e3468.